Back to search resultsSummaryRMgm-932
|
||||||||
 *RMgm-932
*RMgm-932| Successful modification | The parasite was generated by the genetic modification |
| The mutant contains the following genetic modification(s) | Gene disruption |
| Reference (PubMed-PMID number) |
Reference 1 (PMID number) : 24509910 |
| MR4 number | |
| top of page | |
| Parent parasite used to introduce the genetic modification | |
| Rodent Malaria Parasite | P. berghei |
| Parent strain/line | P. berghei ANKA |
| Name parent line/clone | P. berghei ANKA cl15cy1 |
| Other information parent line | A reference wild type clone from the ANKA strain of P. berghei (PubMed: PMID: 17406255). |
| top of page | |
| The mutant parasite was generated by | |
| Name PI/Researcher | T. Annoura; S.M. Khan; C.J. Janse |
| Name Group/Department | Leiden Malaria Research Group |
| Name Institute | Leiden University Medical Center (LUMC) |
| City | Leiden |
| Country | The Netherlands |
| top of page | |
| Name of the mutant parasite | |
| RMgm number | RMgm-932 |
| Principal name | 1309cl1 |
| Alternative name | Δb9-a |
| Standardized name | |
| Is the mutant parasite cloned after genetic modification | Yes |
| top of page | |
| Phenotype | |
| Asexual blood stage | Not different from wild type |
| Gametocyte/Gamete | Not different from wild type |
| Fertilization and ookinete | Not different from wild type |
| Oocyst | Not different from wild type |
| Sporozoite | Not different from wild type |
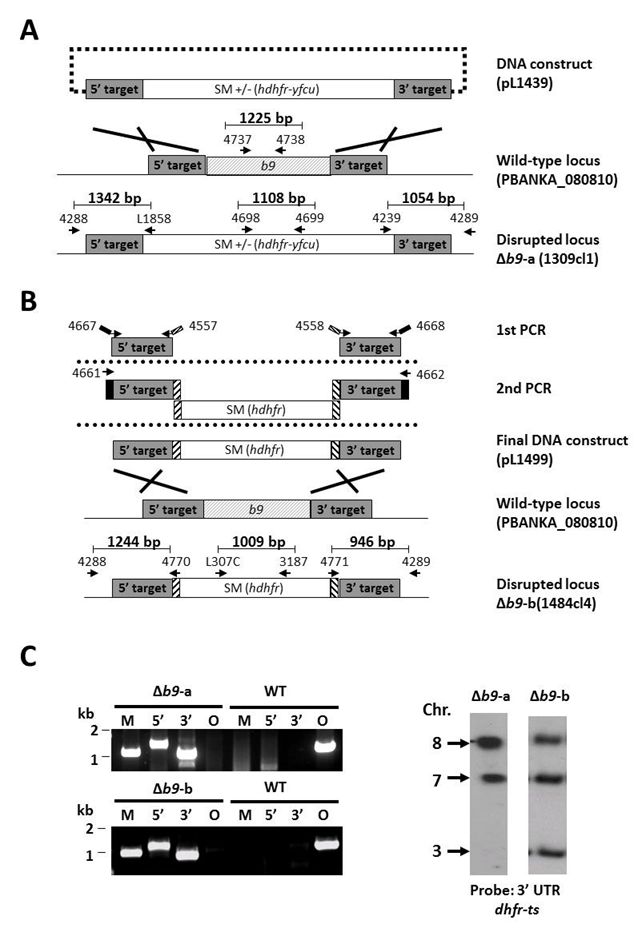
| Liver stage | Normal sporozoite production. Sporozoites showed normal gliding motility and WT-levels of hepatocyte invasion. When Swiss or BALB/c mice were infected by intravenous inoculation of either 1 or 5x104 PbΔb9 sporozoites none of the mice developed blood-stage infections. When C57BL6 mice were infected with a high dose of 5x104 PbΔb9 sporozoites, 10-20% developed a blood-stage infection with a 3-4 days prolonged prepatent period. Immunofluorescence analyses show that PbΔb9 parasites arrest early after invasion of hepatocytes. PbΔb9 sporozoites exhibit normal hepatocyte invasion, but at 24hpi most intra-cellular parasites had disappeared and only a few small parasites could be observed with a size that was similar to 5-10 hpi liver stages. Analysis of PbΔb9 parasites in the liver, using real-time in vivo imaging, confirmed the early growth-arrest observed in cultured hepatocytes. |
| Additional remarks phenotype | Mutant/mutation Supporting Figure S5 Other mutants |
 Disrupted: Mutant parasite with a disrupted gene
Disrupted: Mutant parasite with a disrupted gene| top of page | |||||||||||||||||||||||||
| Details of the target gene | |||||||||||||||||||||||||
| Gene Model of Rodent Parasite | PBANKA_0808100 | ||||||||||||||||||||||||
| Gene Model P. falciparum ortholog | PF3D7_0317100 | ||||||||||||||||||||||||
| Gene product | 6-cysteine protein | ||||||||||||||||||||||||
| Gene product: Alternative name | B9 | ||||||||||||||||||||||||
| top of page | |||||||||||||||||||||||||
| Details of the genetic modification | |||||||||||||||||||||||||
| Inducable system used | No | ||||||||||||||||||||||||
| Additional remarks inducable system | |||||||||||||||||||||||||
| Type of plasmid/construct used | (Linear) plasmid double cross-over | ||||||||||||||||||||||||
| PlasmoGEM (Sanger) construct/vector used | No | ||||||||||||||||||||||||
| Modified PlasmoGEM construct/vector used | No | ||||||||||||||||||||||||
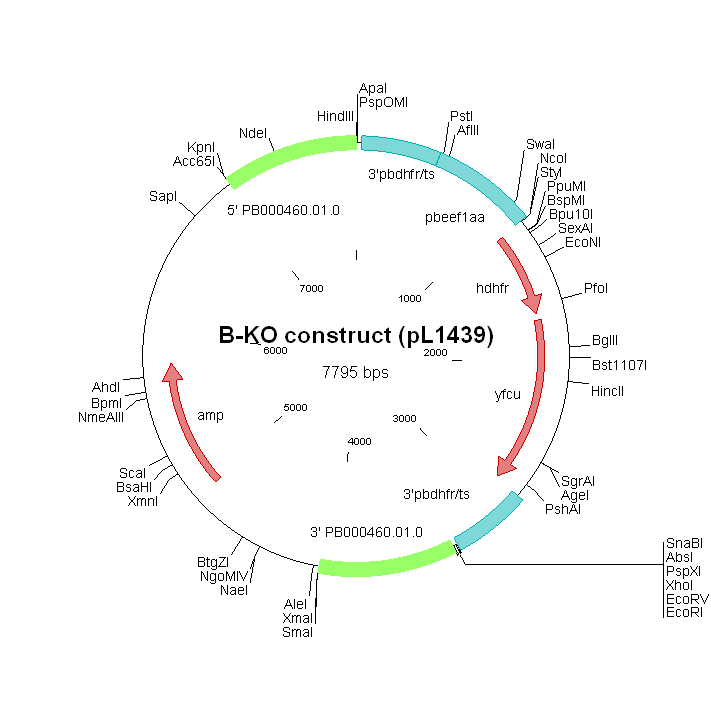
| Plasmid/construct map |
 | ||||||||||||||||||||||||
| Plasmid/construct sequence |
  AGCTTGGGCCCCCGCGGTGGCGGCCGCTCTAGCTTTGATCCCGTTTTTCTTACTTATATA
| ||||||||||||||||||||||||
| Restriction sites to linearize plasmid | |||||||||||||||||||||||||
| Partial or complete disruption of the gene | Complete | ||||||||||||||||||||||||
| Additional remarks partial/complete disruption | |||||||||||||||||||||||||
| Selectable marker used to select the mutant parasite | hdhfr/yfcu | ||||||||||||||||||||||||
| Promoter of the selectable marker | eef1a | ||||||||||||||||||||||||
| Selection (positive) procedure | pyrimethamine | ||||||||||||||||||||||||
| Selection (negative) procedure | No | ||||||||||||||||||||||||
| Additional remarks genetic modification | |||||||||||||||||||||||||
| Additional remarks selection procedure | |||||||||||||||||||||||||
|
Primer information: Primers used for amplification of the target sequences
 Primer information: Primers used for amplification of the target sequences

| |||||||||||||||||||||||||
| top of page | |||||||||||||||||||||||||