| Additional remarks phenotype | Mutant/mutation
The mutant contains a mutated hsp70 locus.
This locus has been mutated by the CRISPR/Cas9 genome editing system through introduction of a double strand break (by Cas9 and a targeting single guide RNA; sgRNA), followed by repair through homologous recombination (see 'Additional information')
Protein (function)
Rodent parasites have several (three, syntenically conserved) hsp70 (related) genes. The orthology of these genes with the (four) hsp70 genes of P. falciparum is not clear
Phenotype
The phenotype has not been analysed in detail. This mutant has been generated to demonstrate gene mutation (nucleotide substitution) by the CRISPR/Cas9 genome editing system (see 'Additional information')
Additional information
The CRISPR/Cas9 genome editing system has been used to delete sera1.
The CRISPR/Cas9 (clustered regularly interspaced short palindromic repeats and Cas9 endonuclease-mediated genome editing) system The CRISPR/Cas9 system was originated from a prokaryotic RNA programmable nuclease that can introduce a double-strand break (DSB) at a specific site on a chromosome through heterologous expression of two components: Cas9 nuclease and a targeting single guide RNA (sgRNA).
Target-specific DSBs introduced by the CRISPR/Cas9 system can be repaired by homologous recombination if a donor template is provided. The CRISPR/Cas9 system has been shown to be highly efficient in other organisms for generating gene knock-in (KI), KO, or allelic replacements.
CRISPR/Cas9-mediated genome editing requires expression of two components: Cas9 nuclease and a targeting single guide RNA (sgRNA), which form a complex to induce a double-strand break (DSB) at the targeted site.
To reduce the size of the plasmid construct and to overcome the problem of limited selectable markers available for P. yoelii, an expression plasmid was constructed that contains the human dihydrofolate reductase (hdhfr)-2A peptide-gfp genes under the P. berghei eef1a (Pbeef1a) promoter and showed bicistronic expression of both genes after introduction into the P. yoelii 17XNL strain.
The viral “ribosome skip” 2A peptide has been shown to coordinate coexpression of two individual genes under a single promoter in P. falciparum
Because Cas9 is a nuclease functioning within the nucleus, two nuclear localization signals (NLSs) were attached to the 5' and 3' of the Cas9 gene to direct the protein to the nucleus.
In mammalian systems, sgRNA is synthesized by RNA polymerase III, and transcription is driven by a U6 small nuclear RNA (snRNA) promoter. By searching the P. yoelii genome database, a U6 snRNA homolog was identified and a 350-base-pair (bp) segment upstream of the transcriptional start site of U6 snRNA was cloned to function as a promoter.
A Cas9-sgRNA plasmid was constructed containing both the hdhfr-2A-SpCas9 and PyU6-sgRNA cassettes with cloning sites for the insertion of donor template sequences (for homologous recombination at target sequences in the genome).
Next a construct was generated to introduce silent mutation of nucleotides into the coding region of the Pyhsp70 gene. First a donor template was generated containing three silent nucleotide substitutions, creating an AatII site into the Pyhsp70 coding region for easy restriction fragment length polymorphism (RFLP) analysis (see Fig. A). One sgRNA was designed to target the site 100 bp upstream of the AatII site (see Fig. 4). To avoid donor DNA digestion by Cas9/sgRNA effector, three nucleotide substitutions were introduced at the binding site of sgRNA in the donor template
(see Fig. B). See Figure below
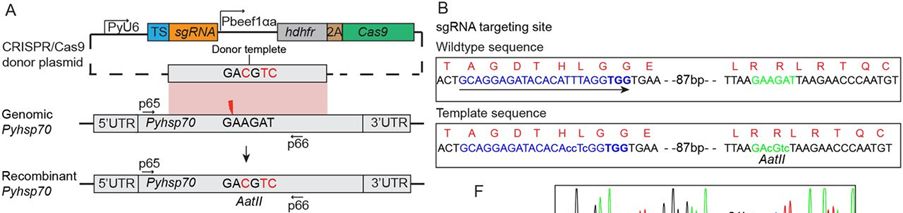
FIG.: Targeted nucleotide replacement in P. yoelii heat shock protein 70 gene (Pyhsp70). (A) Schematic construct for Pyhsp70 nucleotide replacement. The homologous donor template comprises a fragment of Pyhsp70 spanning 340 bp upstream and 620 bp downstream of the Cas9 target site (red thunderbolt). (B) The donor template sequence is identical to the genomic sequence but contains three nucleotide substitutions (green lowercase letters) that create an AatII restriction site for detecting modification by restriction enzyme digestion. In addition, three nucleotides at the sgRNA-binding site in the donor sequence are mutated (blue lowercase letters). The sequence of the protospacer adjacent motif (PAM) is shown in bold type.s.
Considering potential variation in target site accessibility by the Cas9/sgRNA complex, two sgRNAs were designed to target the 3'end of the Pysera1 exon 2, generating plasmids pYC-sera1-sgRNA1 and pYC-sera1-sgRNA2 (see below).
One day after electroporation of the plasmids into the P. yoelii 17XNL strain, parasites were selected with pyrimethamine (Pyr) supplied in drinking water. Pyr-resistant parasites were observed microscopically 5 to 7 days after electroporation.
Other mutants
See RMgm-1096 for gene-tagging mutants that have been generated by this CRISPR/Cas9 system
See RMgm-1095 for gene-deletion (knock-out) mutants that have been generated by this CRISPR/Cas9 system |
 *RMgm-1097
*RMgm-1097 Mutated: Mutant parasite with a mutated gene
Mutated: Mutant parasite with a mutated gene


