Back to search resultsSummaryRMgm-1095
|
||||||||
 *RMgm-1095
*RMgm-1095| Successful modification | The parasite was generated by the genetic modification |
| The mutant contains the following genetic modification(s) | Gene disruption |
| Reference (PubMed-PMID number) |
Reference 1 (PMID number) : 24987097 Reference 2 (PMID number) : 29684399 |
| MR4 number | |
| top of page | |
| Parent parasite used to introduce the genetic modification | |
| Rodent Malaria Parasite | P. yoelii |
| Parent strain/line | P. y. yoelii 17XNL |
| Name parent line/clone | Not applicable |
| Other information parent line | |
| top of page | |
| The mutant parasite was generated by | |
| Name PI/Researcher | Zhang, C; Yuan, J |
| Name Group/Department | State Key Laboratory of Cellular Stress Biology, Innovation Center for Cell Biology, School of Life |
| Name Institute | Xiamen University |
| City | Xiamen, Fujian |
| Country | China |
| top of page | |
| Name of the mutant parasite | |
| RMgm number | RMgm-1095 |
| Principal name | Δsera1 |
| Alternative name | |
| Standardized name | |
| Is the mutant parasite cloned after genetic modification | Yes |
| top of page | |
| Phenotype | |
| Asexual blood stage | Not different from wild type |
| Gametocyte/Gamete | Not different from wild type |
| Fertilization and ookinete | Not different from wild type |
| Oocyst | Not different from wild type |
| Sporozoite | Not different from wild type |
| Liver stage | Not different from wild type |
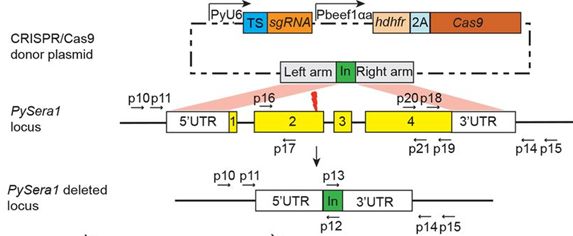
| Additional remarks phenotype | Mutant/mutation In reference PMID 29684399 the authors state: 'To confirm that sera1 is not essential for the parasite, we first disrupted the sera1 gene in the 17XNL parasite using CRISPR/Cas9 methods, and obtained two parasite knockout clones (Δsera1c1 and Δsera1c2) with deletion of the whole coding region. Both Δsera1c1 and Δsera1c2 parasite clones displayed normal progression comparable with wildtype parasite in the life cycle, including asexual and sexual gametocytes stages in mice, mosquito stages, and mouse infectivity (Fig. S1C to G), suggesting functional redundancy of the sera1 gene in the life cycle of P. yoelii. The CRISPR/Cas9 genome editing system has been used to delete sera1. The CRISPR/Cas9 (clustered regularly interspaced short palindromic repeats and Cas9 endonuclease-mediated genome editing) system The CRISPR/Cas9 system was originated from a prokaryotic RNA programmable nuclease that can introduce a double-strand break (DSB) at a specific site on a chromosome through heterologous expression of two components: Cas9 nuclease and a targeting single guide RNA (sgRNA).
|
 Disrupted: Mutant parasite with a disrupted gene
Disrupted: Mutant parasite with a disrupted gene| top of page | |||||||||||||||||||||||||
| Details of the target gene | |||||||||||||||||||||||||
| Gene Model of Rodent Parasite | PY17X_0305700 | ||||||||||||||||||||||||
| Gene Model P. falciparum ortholog | Not available | ||||||||||||||||||||||||
| Gene product | serine repeat antigen 1 | ||||||||||||||||||||||||
| Gene product: Alternative name | SERA1 | ||||||||||||||||||||||||
| top of page | |||||||||||||||||||||||||
| Details of the genetic modification | |||||||||||||||||||||||||
| Inducable system used | No | ||||||||||||||||||||||||
| Additional remarks inducable system | |||||||||||||||||||||||||
| Type of plasmid/construct used | CRISPR/Cas9 construct: integration through double strand break repair | ||||||||||||||||||||||||
| PlasmoGEM (Sanger) construct/vector used | No | ||||||||||||||||||||||||
| Modified PlasmoGEM construct/vector used | No | ||||||||||||||||||||||||
| Plasmid/construct map | |||||||||||||||||||||||||
| Plasmid/construct sequence | |||||||||||||||||||||||||
| Restriction sites to linearize plasmid | |||||||||||||||||||||||||
| Partial or complete disruption of the gene | Complete | ||||||||||||||||||||||||
| Additional remarks partial/complete disruption | |||||||||||||||||||||||||
| Selectable marker used to select the mutant parasite | hdhfr | ||||||||||||||||||||||||
| Promoter of the selectable marker | eef1a | ||||||||||||||||||||||||
| Selection (positive) procedure | pyrimethamine | ||||||||||||||||||||||||
| Selection (negative) procedure | No | ||||||||||||||||||||||||
| Additional remarks genetic modification | To reduce the size of the plasmid construct and to overcome the problem of limited selectable markers available for P. yoelii, an expression plasmid was constructed that contains the human dihydrofolate reductase (hdhfr)-2A peptide-gfp genes under the P. berghei eef1a (Pbeef1a) promoter and showed bicistronic expression of both genes after introduction into the P. yoelii 17XNL strain. The viral “ribosome skip” 2A peptide has been shown to coordinate coexpression of two individual genes under a single promoter in P. falciparum Because Cas9 is a nuclease functioning within the nucleus, two nuclear localization signals (NLSs) were attached to the 5' and 3' of the Cas9 gene to direct the protein to the nucleus. In mammalian systems, sgRNA is synthesized by RNA polymerase III, and transcription is driven by a U6 small nuclear RNA (snRNA) promoter. By searching the P. yoelii genome database, a U6 snRNA homolog was identified and a 350-base-pair (bp) segment upstream of the transcriptional start site of U6 snRNA was cloned to function as a promoter. A Cas9-sgRNA plasmid was constructed containing both the hdhfr-2A-SpCas9 and PyU6-sgRNA cassettes with cloning sites for the insertion of donor template sequences (for homologous recombination at target sequences in the genome). Next the plasmid pYC-Pysera1 was constructed containing a 46-bp tag DNA (for PCR primers) flanked by two homologous regions of Pysera1 (0.7 kb of the 5'-flanking region and 0.8 kb of the 3'-flanking region) Considering potential variation in target site accessibility by the Cas9/sgRNA complex, two sgRNAs were designed to target the 3'end of the Pysera1 exon 2, generating plasmids pYC-sera1-sgRNA1 and pYC-sera1-sgRNA2. The plasmid containing sgRNA and Cas9 (and 3'/5'-sera homology regions) contains a hdhfr selection cassette. This cassette is not integrated into the genome. Sequence of the sgRNA: sgRNA 1: GTACCGGGAAACTCTGATCAAGG (genomic target sequence); GTACCGGGAAACTCTGATCA (sgRNA targetr sequence) sgRNA 2: GCTAGCGACTGTTTCTTACATGG (genomic target sequence); GCTAGCGACTGTTTCTTACA (sgRNA targetr sequence) See below for primer sequences used to amplify the targeting regions of sera1 | ||||||||||||||||||||||||
| Additional remarks selection procedure | |||||||||||||||||||||||||
|
Primer information: Primers used for amplification of the target sequences
 Primer information: Primers used for amplification of the target sequences

| |||||||||||||||||||||||||
| top of page | |||||||||||||||||||||||||